
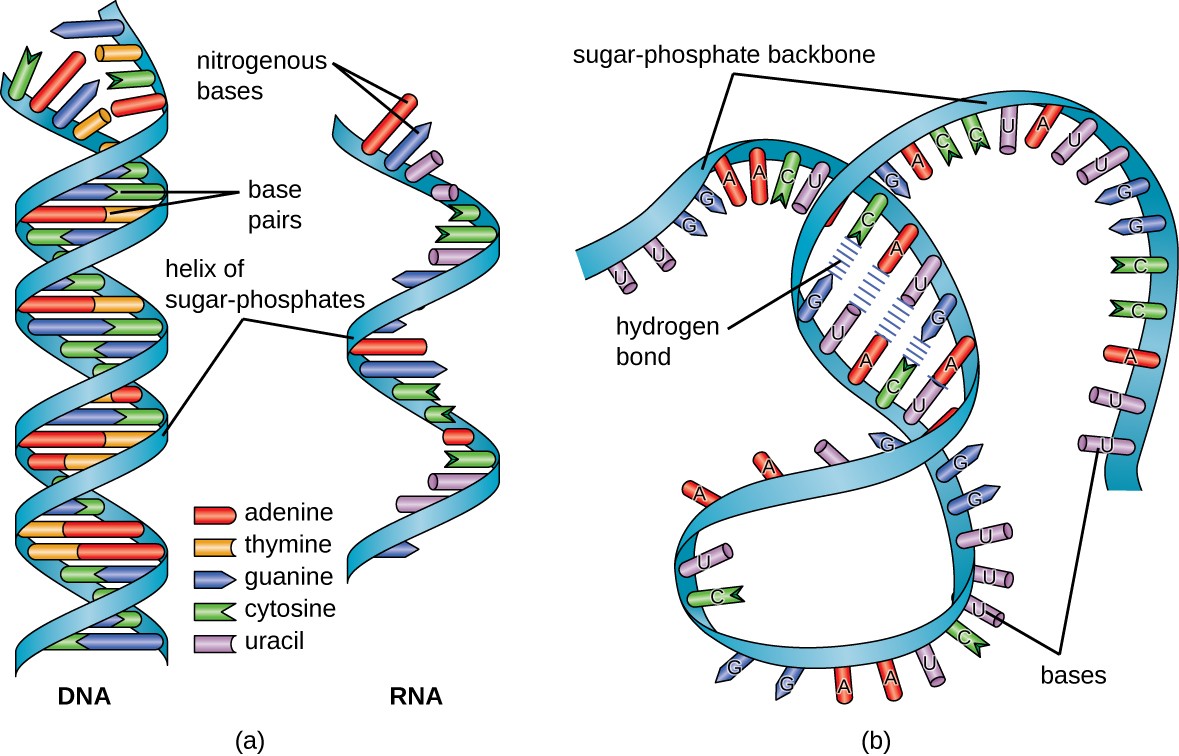
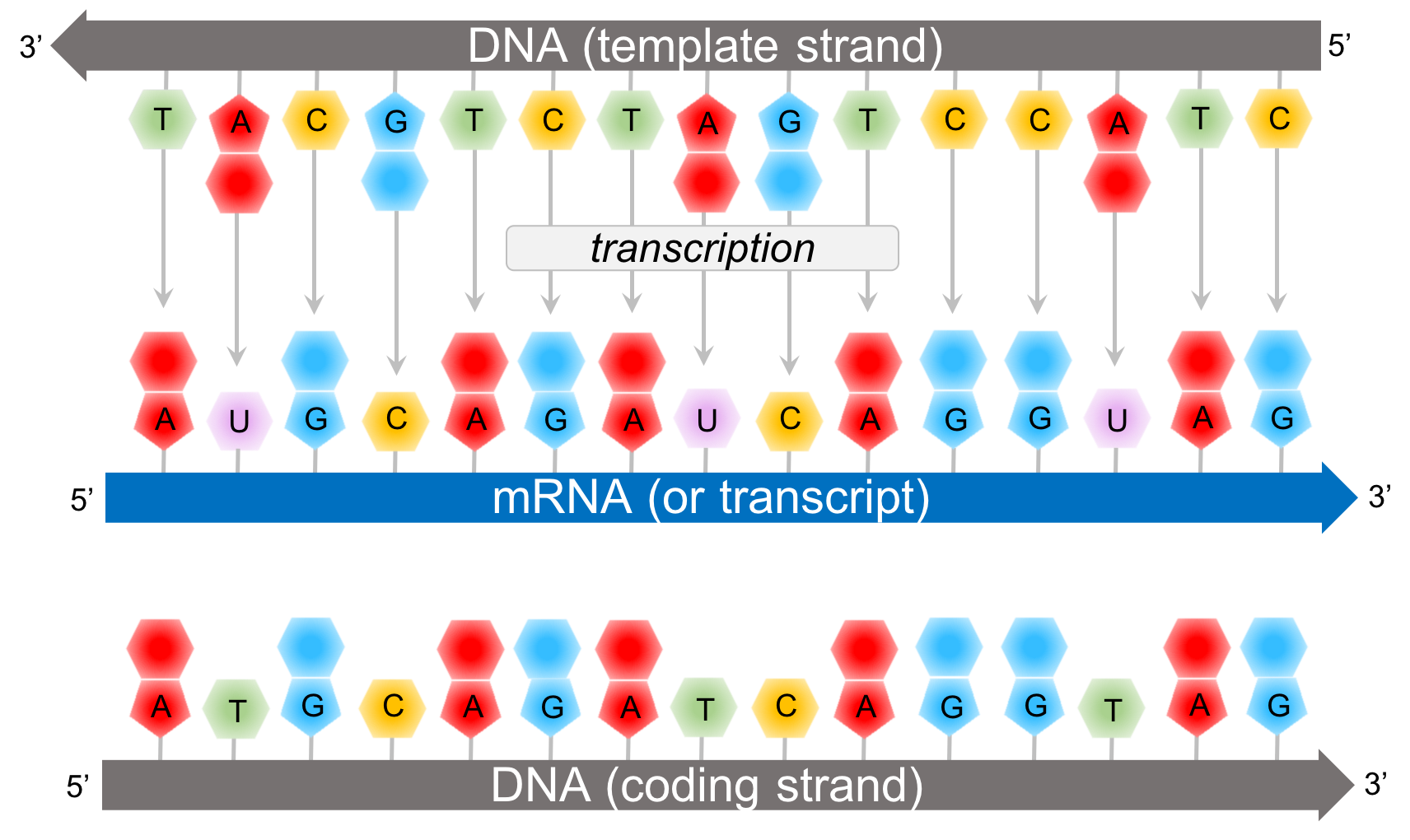
The adaptors contain constant regions that serve as primer-binding sites for subsequent PCR amplification, and these are usually asymmetric so that the “strandedness” of the template is preserved. Oligonucleotide sequences known as adaptors are then ligated onto the DNA fragments. Because RNA is inherently prone to degradation, it is first reverse transcribed to double-stranded DNA. MicroRNAs, which are 22-nucleotide regulatory RNAs, can be selectively isolated for sequencing based on their size. For example, protein-coding messenger RNAs, or mRNAs, can be captured with “oligo-dT”-short stretches of deoxy-T nucleotides that bind to the sequence of A bases known as a poly-A tail at the end of these transcripts. To facilitate recovery of other types of transcripts, rRNA is typically removed prior to sequencing by hybridizing the sample to complementary oligonucleotides attached to magnetic beads, and using a magnet to separate the rRNA from the rest of the sample.Īlternatively, a specific population of RNA can be selected for sequencing. Most RNA in cells is ribosomal RNA, or rRNA, the central component of the cell’s protein-production machinery. Transcriptome sequencing requires isolating a population of transcripts whose levels are to be measured. First, let’s review some principles behind RNA-seq.


 0 kommentar(er)
0 kommentar(er)
